When you render, R Markdown 1. Runs the R code, embeds results and text into.md file with knitr 2. Then converts the.md file into the finished format with pandoc Set a document’s default output format in the YAML header:- output: htmldocument - # Body output value creates htmldocument html pdfdocument pdf (requires Tex ). Stargazer and R Markdown to LaTeX If you're using LaTeX, then use the Sweave-like knitr opions here instead. The `results='asis'` means that knitr won't do any processing on the output, so when you run this it produces a bunch of gobbledygook.
R Markdown is an open-source tool for producing reproducible reports in R. It enables you to keep all of your code, results, plots, and writing in one place. R Markdown is particularly useful when you are producing a document for an audience that is interested in the results from your analysis, but not your code. Rmarkdownパッケージを使用してレポートを作成しようとしていますが、残念ながら、フィールドレポートではMS Word文書として提出されることが多いようです。だから私はいつもの力に頼ることはできません.
Overview
QuestionsHow can I integrate software and reports?
Value of reproducible reports
Basics of Markdown
R code chunks
Chunk options
Inline R code
Other output formats
Data analysis reports
Data analysts tend to write a lot of reports, describing theiranalyses and results, for their collaborators or to document theirwork for future reference.
When I was first starting out, I’d write an R script with all of mywork, and would just send an email to my collaborator, describing theresults and attaching various graphs. In discussing the results, therewould often be confusion about which graph was which.
I moved to writing formal reports, with Word or LaTeX, but I’d have tospend a lot of time getting the figures to look right. Mostly, theconcern is about page breaks.
Everything is easier now that I create a web page (as an htmlfile). It can be one long stream, so I can use tall figures thatwouldn’t ordinary fit on one page. Scrolling is your friend.
Literate programming
Ideally, such analysis reports are reproducible documents: If anerror is discovered, or if some additional subjects are added to thedata, you can just re-compile the report and get the new or correctedresults (versus having to reconstruct figures, paste them intoa Word document, and further hand-edit various detailed results).
The key tool for R is knitr, which allowsyou to create a document that is a mixture of text and some chunks ofcode. When the document is processed by knitr, chunks of R code willbe executed, and graphs or other results inserted.
This sort of idea has been called “literate programming”.
knitr allows you to mix basically any sort of text with any sort ofcode, but we recommend that you use R Markdown, which mixes Markdownwith R. Markdown is a light-weight mark-up language for creating webpages.
Creating an R Markdown file
Within R Studio, click File → New File → R Markdown andyou’ll get a dialog box like this:
You can stick with the default (HTML output), but give it a title.
Basic components of R Markdown
The initial chunk of text contains instructions for R: you give thething a title, author, and date, and tell it that you’re going to wantto produce html output (in other words, a web page).
You can delete any of those fields if you don’t want themincluded. The double-quotes aren’t strictly necessary in this case.They’re mostly needed if you want to include a colon in the title.
RStudio creates the document with some example text to get youstarted. Note below that there are chunks like
These are chunks of R code that will be executed by knitr and replacedby their results. More on this later.
Also note the web address that’s put between angle brackets (< >) aswell as the double-asterisks in **Knit**. This isMarkdown.
Markdown
Markdown is a system for writing web pages by marking up the text muchas you would in an email rather than writing html code. The marked-uptext gets converted to html, replacing the marks with the properhtml code.
For now, let’s delete all of the stuff that’s there and write a bit ofmarkdown.
You make things bold using two asterisks, like this: **bold**,and you make things italics by using underscores, like this:_italics_.
Knitr R Markdown From Command Line
You can make a bulleted list by writing a list with hyphens orasterisks, like this:
or like this:
Each will appear as:
- bold with double-asterisks
- italics with underscores
- code-type font with backticks
(I prefer hyphens over asterisks, myself.)
You can make a numbered list by just using numbers. You can use thesame number over and over if you want:
This will appear as:
- bold with double-asterisks
- italics with underscores
- code-type font with backticks
You can make section headers of different sizes by initiating a linewith some number of # symbols:
You compile the R Markdown document to an html webpage by clickingthe “Knit HTML” in the upper-left. And note the little question marknext to it; click the question mark and you’ll get a “Markdown QuickReference” (with the Markdown syntax) as well to the RStudiodocumentation on R Markdown.
Challenge
Create a new R Markdown document. Delete all of the R code chunksand write a bit of Markdown (some sections, some italicizedtext, and an itemized list).
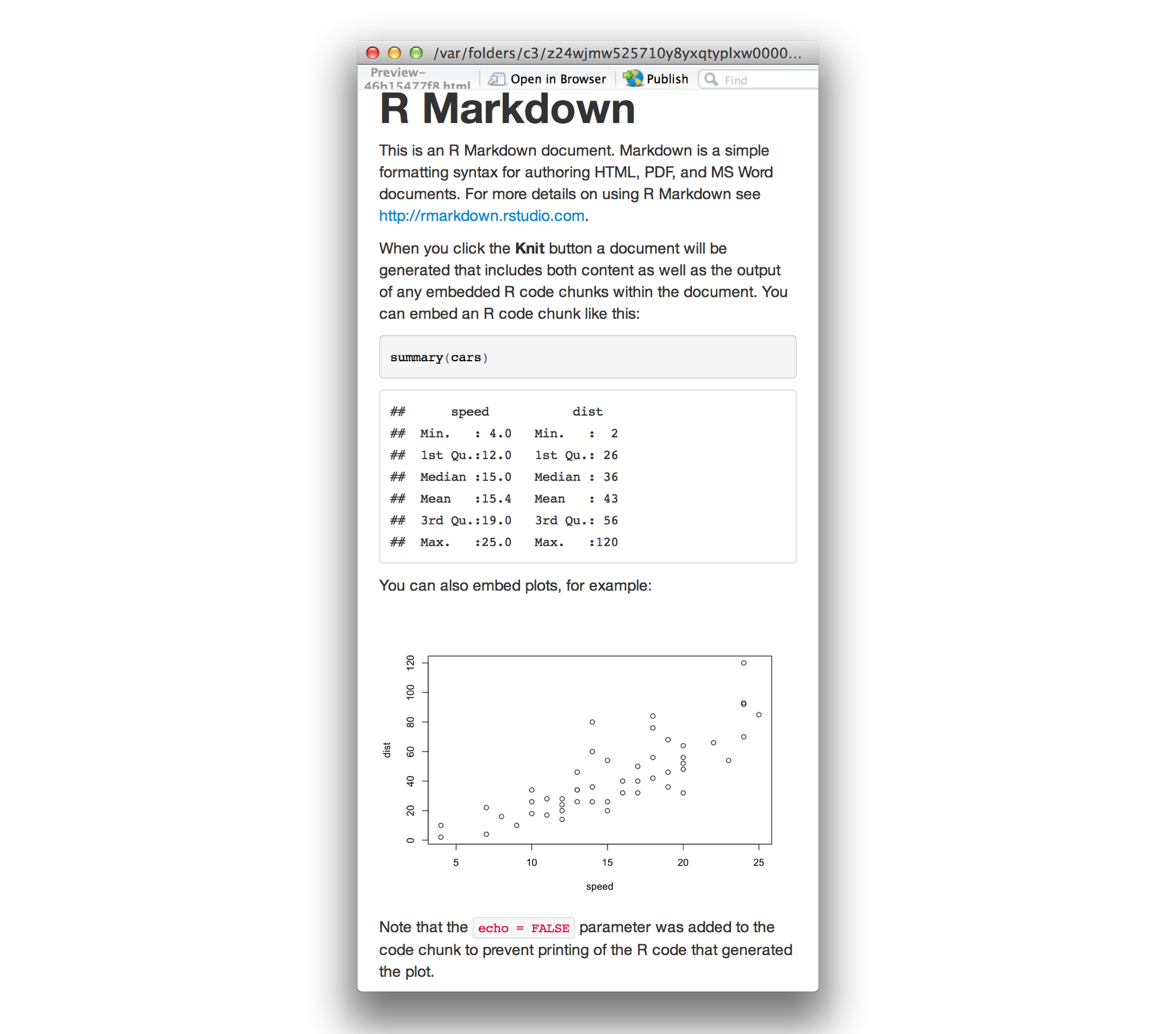
Convert the document to a webpage.
A bit more Markdown
You can make a hyperlink like this:[text to show](http://the-web-page.com).
You can include an image file like this: 
You can do subscripts (e.g., F~2~) with F~2 and superscripts (e.g.,F^2^) with F^2^.
If you know how to write equations inLaTeX, you’ll be glad to know thatyou can use $ $ and $$ $$ to insert math equations, like$E = mc^2$ and
R code chunks
Markdown is interesting and useful, but the real power comes frommixing markdown with chunks of R code. This is R Markdown. Whenprocessed, the R code will be executed; if they produce figures, thefigures will be inserted in the final document.
The main code chunks look like this:
That is, you place a chunk of R code between ```{r chunk_name}and ```. It’s a good idea to give each chunka name, as they will help you to fix errors and, if any graphs areproduced, the file names are based on the name of the code chunk thatproduced them.
Challenge
Add code chunks to
- Load the ggplot2 package
- Read the gapminder data
- Create a plot
How things get compiled
When you press the “Knit HTML” button, the R Markdown document isprocessed by knitr and a plain Markdowndocument is produced (as well as, potentially, a set of figure files): the R code is executedand replaced by both the input and the output; if figures areproduced, links to those figures are included.
The Markdown and figure documents are then processed by the toolpandoc, which converts the Markdown file into anhtml file, with the figures embedded.
Chunk options
There are a variety of options to affect how the code chunks aretreated.
- Use
echo=FALSEto avoid having the code itself shown. - Use
results='hide'to avoid having any results printed. - Use
eval=FALSEto have the code shown but not evaluated. - Use
warning=FALSEandmessage=FALSEto hide any warnings ormessages produced. - Use
fig.heightandfig.widthto control the size of the figuresproduced (in inches).
So you might write:
Often there will be particular options that you’ll want to userepeatedly; for this, you can set global chunk options, like so:
The fig.path option defines where the figures will be saved. The /here is really important; without it, the figures would be saved inthe standard place but just with names that being with Figs.
If you have multiple R Markdown files in a common directory, you mightwant to use fig.path to define separate prefixes for the figure filenames, like fig.path='Figs/cleaning-' and fig.path='Figs/analysis-'.
Challenge
Use chunk options to control the size of a figure and to hide thecode.
Inline R code
You can make every number in your report reproducible. Use`r and ` for an in-line code chunk,like so: `r round(some_value, 2)`. The code will beexecuted and replaced with the value of the result.
Don’t let these in-line chunks get split across lines.
Perhaps precede the paragraph with a larger code chunk that doescalculations and defines things, with include=FALSE for that largerchunk (which is the same as echo=FALSE and results='hide').
I’m very particular about rounding in such situations. I may want2.0, but round(2.03, 1) will give just 2.
Themyroundfunction in my R/broman package handlesthis.
Challenge
Try out a bit of in-line R code.
Other output options


You can also convert R Markdown to a PDF or a Word document. Click thelittle triangle next to the “Knit HTML” button to get a drop-downmenu. Or you could put pdf_document or word_document in the headerof the file.
Tip: Creating PDF documents
Creating .pdf documents may require installation of some extra software. Ifrequired this is detailed in an error message.
Tex for windows is available here.
Tex for mac is available here.
Resources
- Dynamic Documents with R and knitr (book)
Key Points
Mix reporting written in R Markdown with software written in R.
Specify chunk options to control formatting.
Use
knitrto convert these documents into PDF and other formats.
- This post was updated with advanced tips (at the bottom) 2/12/2016
For scientific projects we often using knitr and R markdown in RStudio to share results and code either in HTML or PDF format. These are great tools for reproducible research. We find, however, that keeping all of our R code in an R markdown (.Rmd) file creates two problems. First, you need to completely re-run the report to evaluate new bits of R code and secondly the .Rmd file can become unwieldy very quickly. Fortunately, there is an easy way to make use of external code.
1. Create your external R script
You would create an R script as normal and, within the R script, you create ‘chunks’ using the ## @knitr syntax. So, in this example, we create a new R script entitled example.R with two chunks (variablesXY and plotXY) that looks like this:
2. Create your R markdown script and refer to the external R script
Now you can create your R markdown (.Rmd) file. In order to read your external file you use the function read_chunk and then you can reference individual chunks using the <<chunkname>> syntax. So, continuing with our example, we can create a new file called myreport.Rmd, read the external file and reference/run the code chunks:
3. Alternative syntax
R Markdown Knitr Options
Although we usually use the syntax above which allows us to easily re-use chunks and flexibly name the R markdown chunks, there is an alternative syntax you can use. You can move all of the R code to the chunks in the external file and refer to those chunks in the R markdown chunk headers. For example, to produce exactly the same output as that above you can use the following for the example.R:
And then the myreport.Rmd can be simplified to look like:
Knitr Kable R Markdown
In either case, the output would look like this:
Advanced tips:
Run and show external code but leave off the comments
This was a tip provided by Bob Rudis. My external code often has comments that I don't want included in my final report but including code and excluding comments is tricky. To do this you add a couple of arguments to your code chunk, one is tidy=TRUE which tells R to tidy up the code (but leaves comments intact). The second is an argument to tidy.opts setting comment=FALSE. To a certain extent this is covered in Yihui's post on chunk options but the various setting available in tidy.opts are not spelled out there.
Run or show all code from an external script
I occasionally have a situation where I want to echo the code from a script but I don't want to run it. The simplest way to do this would be to copy and paste the code into a code chunk (and set eval=FALSE) but, of course, you would need to re-cut and paste if your code changes. An alternative to this, explained by Yihui at the bottom of this page, would be to use the code argument and readLines. So, as an example, to echo the code (but don't run it) from an external script called myscript.R in your Rmarkdown you can use:
